News
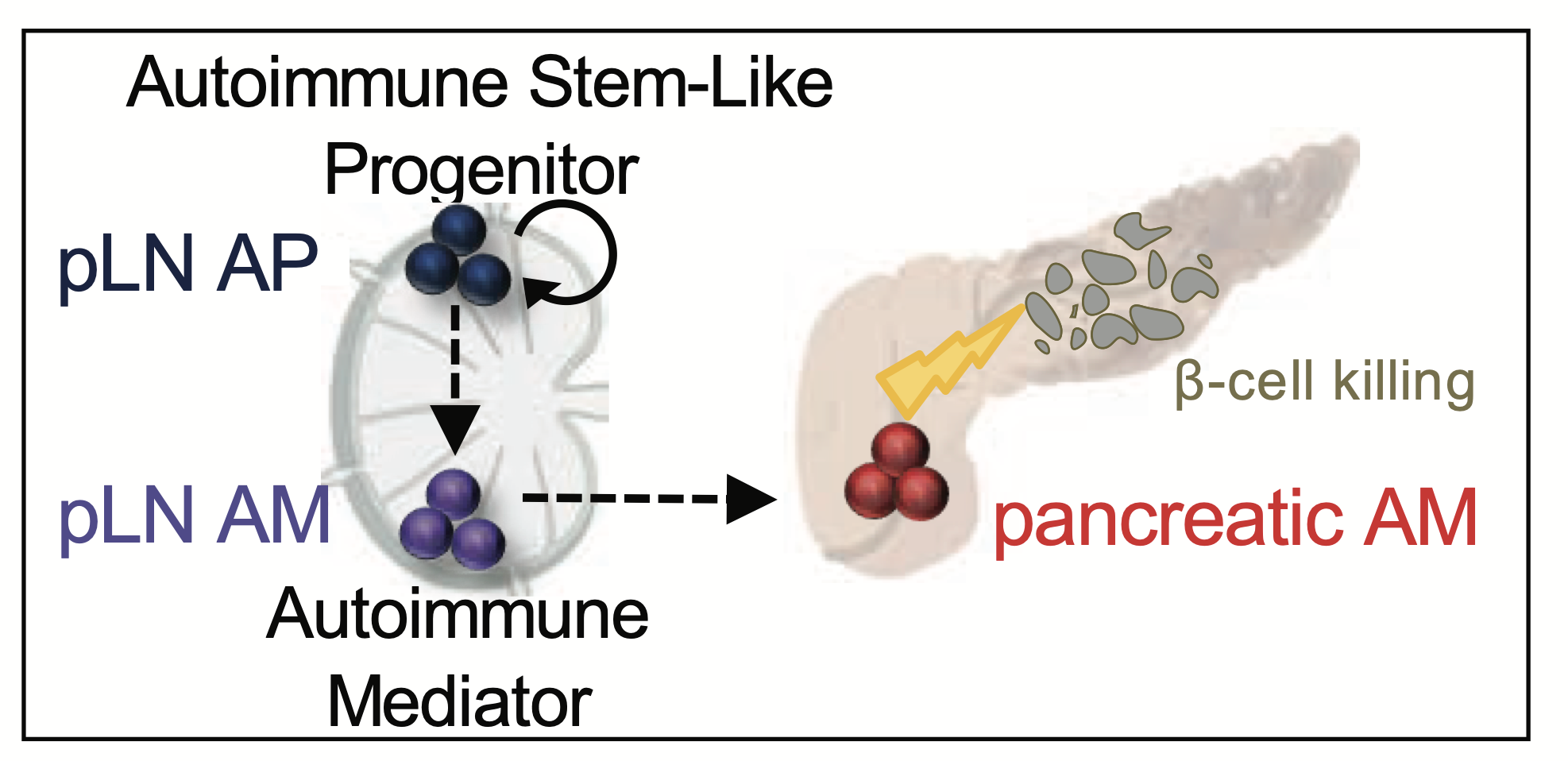
- [January 2026] New Nature publication in collaboration with Tuomas Tammela describing lineage tracing of high plasticity cell state (HPCS) in lung adenocarcinoma demonstrating its role in transitioning between cell states and contributing to treatment resistance
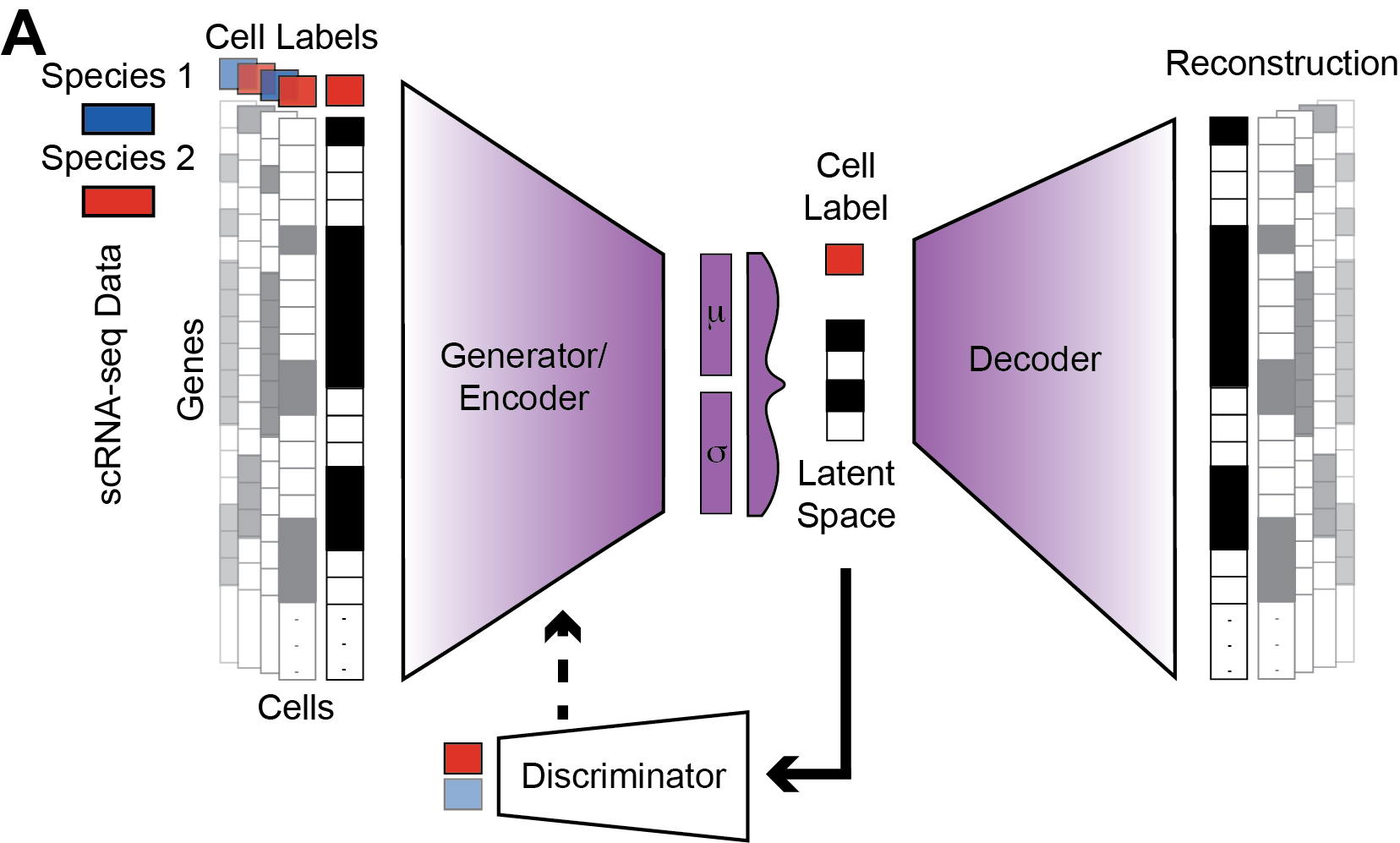
- [December 2025] The latest paper from our lab is out in Cancer Research . This is a deep-learning model for cross species scRNA-seq integration termed scVital. Led by our group's recent graduate student Jonathan Rub and in close collaboration with Tuomas Tammela. Source code is available in Github .
- [April 2025] Our recent Aging Cell paper, led by Chao Zhang in collaboration with the Studer lab. We developed a cellular aging score that was used to identify new approaches for in vitro age modulation. Part of our ongoing collaboration with Lorenz Studer and his group developing new approaches of stem cell models of neurodegenerative diseases.
- [December 2024] A new prepring from our lab is posted. This study, led by Fayzan Chaudhry, is an identification of plasma proteins biomarkers of Parkinson's disease using large public biomedical database
Open Positions
We are looking for graduate students in the area of computational
genomics. Trainees will be working on:
- Developing new machine learning algorithms for the analysis of single cell data and spatial genomics.
- Participating and leading multi-desciplinary projects with collaborators in immunology (autoimmune, tumor immunology), stem cell and cancer genetics.
Interested graduate students should apply through Weill Cornell graduate school to the CBM or PBSB programs.